- Dmitrii E. Makarov, The effect of a mechanical force on quantum reaction rate: Quantum Bell formula, J. Chem. Phys. 135 (2011), 194112, link
- Sai Sriharsha M. Konda, Johnathan N. Brantley, Christopher W. Bielawski, and Dmitrii E. Makarov, Chemical reactions modulated by mechanical stress: Extended Bell theory, J. Chem. Phys. 135, 164103 (2011) , link
- Dmitrii E. Makarov, Individual Proteins Under Mechanical Stress: Lessons from Theory and Computer Simulations, In Single-Molecule Studies of Proteins, A. Oberhauser (Editor), Springer, 2013, link to the book
- Sai Sriharsha M. Konda, Johnathan N. Brantley, Bibin T. Varghese, Kelly M.Wiggins, Christopher W. Bielawski, and Dmitrii E. Makarov, Molecular Catch Bonds and the anti-Hammond Effect in Polymer Mechanochemistry, JACS, 135, 12722-12729 (2013), link
- Sai Sriharsha M. Konda, Stanislav M. Avdoshenko, and Dmitrii E. Makarov, Exploring the topography of the stress-modified energy landscapes of mechanosensitive molecules, J. Chem. Phys. 140, 104114 (2014), link
- Stanislav M. Avdoshenko, Sai Sriharsha M. Konda, and Dmitrii E. Makarov, On the calculation of internal forces in mechanically stressed polyatomic molecules, J. Chem. Phys. 141, 134115 (2014), link
Timescales in reaction dynamics
- Chun-Yaung Lu, Dmitrii E. Makarov and Graeme Henkelman, Kappa-dynamics-An exact method for accelerating rare event classical molecular dynamics, J. Chem. Phys. 133 (2010) 201101, link
- Srabanti Chaudhury and Dmitrii E. Makarov, A harmonic transition state approximation for the duration of reactive events in complex molecular rearrangements, J. Chem. Phys. 133 (2010) 034118, link
- Ryan R. Cheng an Dmitrii E. Makarov, Failure of one-dimensional Smoluchowski diffusion models to describe the duration of conformational rearrangements in floppy, diffusive molecular systems: A case study of polymer cyclization, J. Chem. Phys., 134 (2011) 085104, link
- Alex T. Hawk and Dmitrii E. Makarov, Milestoning with transition memory, J. Chem. Phys. 135 (2011), 224109, link
Single-molecule dynamics: theory, interpretation and data analysis
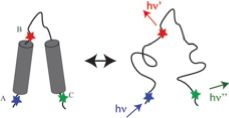
- Z.S. Wang and D.E. Makarov, Nanosecond dynamics of single polypeptide molecules revealed by photoemission statistics of fluorescence resonance energy transfer: A theoretical study, J. Phys. Chem. B 107 (2003) 5617-5622. Link to text
- Evan R. McCarney, James H. Werner, Ingo Ruczinski, Dmitrii E. Makarov, Richard A. Keller, Peter M. Goodwin, Kevin W. Plaxco, Site-specific deviations from random coil dimensions in a highly denatured protein; A single molecule study. Journal of Molecular Biology, 352 (2005) 672-682.
- Christina L. Ting and Dmitrii E. Makarov, Two-dimensional fluorescence resonance energy transfer as a probe for protein folding: A theoretical study, J. Chem. Phys. 128 (2008) 115102. link
- Dmitrii E. Makarov and Kevin W. Plaxco, Measuring the dimensions of unfolded biopolymers using FRET: The effect of polymer chain dynamics on the observed FRET efficiency, Journal of Chemical Physics, 131 (2009), 085105.link
- J. Nick Taylor, Dmitrii E. Makarov, and Christy F. Landes, Denoising Single-Molecule FRET Trajectories with Wavelets and Bayesian Inference, Biophysical Journal 98 (2010), 164-173. link
- Dmitrii E. Makarov, Spatio-temporal correlations in denatured proteins: the dependence of FRET-derived reconfiguration times on the location of the FRET probes, Journal of Chemical Physics 132 (2010) 035104. link
- Andrea Soranno, Brigitte Buchli, Daniel Nettels, Sonja Müller-Späth, Ryan R. Cheng, Shawn H. Pfeil, Armin Hoffmann, Everett A. Lipman, Dmitrii E. Makarov, Benjamin Schuler, Quantifying internal friction in unfolded and intrinsically disordered proteins with single molecule spectroscopy, PNAS (2012) link
- Ignacia Echeverria, Dmitrii E. Makarov, and Garegin A. Papoian, Concerted Dihedral Rotations Give Rise to Internal Friction in Unfolded Proteins, JACS, DOI: 10.1021/ja503069k link
Dynamics of topologically and spatially constrained polymers: loops, knots, and surface-confined polymers
- Lei Huang and Dmitrii E. Makarov, Langevin dynamics simulations of the diffusion of molecular knots in tensioned polymer chains, J. Phys. Chem. A 111(2007) 10338-10344 link
- Serdal Kirmizialtin and Dmitrii E. Makarov, Simulations of the untying of molecular friction knots between individual polymer strands, J. Chem. Phys. 128 (2008) 094901 link
- Takanori Uzawa, Ryan R. Cheng, Kevin J Cash, Dmitrii E. Makarov and Kevin W Plaxco, The Length and Viscosity Dependence of End-to-end Collision Rates in Single-stranded DNA, Biophysical Journal 97 (2009), 205-210. link
- Ryan R. Cheng, Takanori Uzawa, Kevin W Plaxco, and Dmitrii E. Makarov, The rate of intramolecular loop formation in DNA and polypeptides: The absence of the diffusion-controlled limit and fractional power law viscosity dependence, Journal of Physical Chemistry B 113 (2009), 14026-14034. link
- Ryan R. Cheng and Dmitrii E. Makarov, End-to-surface reaction dynamics of a single surface-attached DNA or polypeptide, J. Phys. Chem. B 114 (2010), 3321-3329.link
- Ryan R. Cheng, Takanori Uzawa, Kevin W. Plaxco, and Dmitrii E. Makarov, Universality in the Timescales of Internal Loop Formation in Unfolded Proteins and Single-Stranded Oligonucleotides, Biophysical Journal, 99 (2010), 3959-3968. link
- Takanori Uzawa, Ryan R. Cheng, Ryan J. White, Dmitrii E. Makarov, and Kevin W. Plaxco, A Mechanistic Study of Electron Transfer from the Distal Termini of Electrode-Bound, Single-Stranded DNAs, JACS 132 (2010), 16120-16126, link
- Herschel M. Watkins, Alexis Vallée-Bélisle, Francesco Ricci, Dmitrii E. Makarov, and Kevin W. Plaxco, Entropic and Electrostatic Effects on the Folding Free Energy of a Surface-Attached Biomolecule: An Experimental and Theoretical Study, JACS (2012), 134(4) 2120-2126. link
- Takanori Uzawa, Takashi Isoshima, Yoshihiro Ito, Koichiro Ishimori, Dmitrii E. Makarov, and Kevin W. Plaxco, Sequence and Temperature Dependence of the End-to-End Collision Dynamics of Single-Stranded DNA, Biophys. Journal 104 (2013), 2485-2492, link
Protein translocation
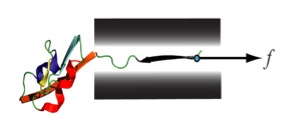
- Serdal Krimizialtin, Venkat Ganesan, and Dmitrii E. Makarov, Translocation of a beta-hairpin-forming peptide through a cylindrical tunnel, J. Chem. Phys. 121 (2004) 10268. link
- Lei Huang, Serdal Kirmizialtin, and Dmitrii E. Makarov, Computer simulations of the translocation and unfolding of a protein pulled mechanically through a pore, J. Chem. Phys. 123 (2005) 124903 link
- Serdal Kirmizialtin, Lei Huang, and Dmitrii E. Makarov, Computer simulations of protein translocation, Physica Status Solidi (b) 243 (2006) 2038-2047 link
- Carl P. Goodrich, Serdal Kirmizialtin, Beatrice M. Huyghues-Despointes, Aiping Zhu, J. Martin Scholtz, Dmitrii E. Makarov and Liviu Movileanu, Single-molecule Electrophoresis of b-Hairpin-forming Peptides by Electrical Recordings and Langevin Dynamics Simulations, 111 (2007) 3332-3335 link
- Lei Huang and Dmitrii E. Makarov, The rate constant of polymer reversal inside a pore, J. Chem. Phys. 128 (2008) 114903 link
- Lei Huang and Dmitrii E. Makarov, Translocation of a knotted polypeptide through a pore, J. Chem. Phys. 129 (2008), 121107. link
- Dmitrii E. Makarov, Computer simulations and theory of protein translocation, Accounts of Chemical Research, 42 (2009) 281-289. link
- Dmitrii E. Makarov, Computational and Theoretical Insights into Protein Translocation, Protein & Peptide Letters, 21 (2014), 217-226
Single protein molecules under mechanical tension

- D.E. Makarov, P.K. Hansma, and H. Metiu, Kinetic Monte Carlo simulation of titin unfolding, J. Chem. Phys. 114 (2001) 9663. Link to the article text
- D.E. Makarov, Z. Wang, J. Thompson, H.G. Hansma, On the interpretation of force extension curves of single protein molecules, J. Chem. Phys. 116 (2002) 7760. Link
- D.E. Makarov and G.J. Rodin, Configurational entropy and mechanical properties of cross-linked polymer chains, Implications for protein and RNA folding, Phys. Rev. E 66 (2002) 011908 link to text
- Nathan Becker, Emin Oroudjev, Stephanie Mutz, Jason Cleveland, Paul K. Hansma, Cheryl Y. Hayashi, Dmitrii E. Makarov, and Helen Hansma, Molecular Nanosprings in Spider Capture-Silk Threads, Nature Materials 2 (2003) 278
- Kilho Eom, Pai-Chi Li, Dmitrii E. Makarov, and Gregory J. Rodin, Relationship between the mechanical properties and topology of cross-linked polymer molecules: Parallel strands maximize the strength of model polymers and protein domains, J. Phys. Chem. B 107 (2003) 8730. Link to text published online
- Pai-Chi Li and Dmitrii E. Makarov, Theoretical studies of the mechanical unfolding of the muscle protein titin: Bridging the time-scale gap between simulation and experiment, J. Chem. Phys 119 (2003) 9260. link to text
- Pai-Chi Li and Dmitrii E. Makarov, Ubiquitin-like protein domains show high resistance to mechanical unfolding similar to that of the I27 domain in titin: Evidence from simulations, J. Phys. Chem. B. 108 (2004) 745-749. link
- Pai-Chi Li and Dmitrii E. Makarov, Simulation of the mechanical unfolding of ubiquitin: Probing different unfolding reaction coordinates by changing the pulling geometry, J. Chem. Phys. 121 (2004) 4826. link
- Kilho Eom, Dmitrii E. Makarov, and Gregory J. Rodin, Theoretical studies of the kinetics of mechanical unfolding of cross-linked polymer chains and their implications for single molecule pulling experiments, Phys. Rev. E 71, 021904 (2005).link
- Serdal Kirmizialtin, Lei Huang, and Dmitrii E. Makarov, Topography of the free energy landscape probed via mechanical unfolding of proteins, J. Chem. Phys. 122 (2005) 234915 link
- Pai-Chi Li, Lei Huang, and Dmitrii E. Makarov, Mechanical unfolding of segment-swapped protein G dimer: Results from replica exchange molecular dynamics simulations., J. Phys. Chem B. 110 (2006) 14469-14474 link
- Dmitrii E. Makarov, New and Notable: unraveling individual molecules by mechanical forces: Theory meets experiment, Biophysical Journal, 92 (2007) 4135-4136 link (This is a commentary on this paper)
- Dmitrii E. Makarov, A theoretical model for the mechanical unfolding of repeat proteins, Biophysical Journal 96 (2009) 2160 link
- Dmitrii E. Makarov, Individual Proteins Under Mechanical Stress: Lessons from Theory and Computer Simulations, In Single-Molecule Studies of Proteins, A. Oberhauser (Editor), Springer, 2013, link to the book
Protein dynamics and folding
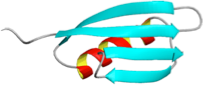
- D.E. Makarov, C. Keller, K.W. Plaxco, and H. Metiu, How the folding rate constant of simple, single-domain proteins depends on the number of native contacts, Proc. Natl. Acad. Sci. USA 99 (2002) 3535 link to the article text
- D.E. Makarov and H. Metiu, A model for the kinetics of protein folding: Kinetic Monte Carlo simulations and analytical results, J. Chem. Phys. 116 (2002) 5205 link to the article text
- Z. S. Wang and D. E. Makarov, Rate of intramolecular contact formation in peptides: The loop length dependence, J. Chem. Phys. 117 (2002) 4591 link to text
- D.E. Makarov and G.J. Rodin, Configurational entropy and mechanical properties of cross-linked polymer chains, Implications for protein and RNA folding, Phys. Rev. E 66 (2002) 011908 link to text
- D.E. Makarov and K.W. Plaxco, The Topomer Search Model: A simple, quantitative theory of two-state protein folding kinetics, Protein Science 12 (2003) 17-26 link
- Serdal Krimizialtin, Venkat Ganesan, and Dmitrii E. Makarov, Translocation of a beta-hairpin-forming peptide through a cylindrical tunnel, J. Chem. Phys. 121 (2004) 10268. link
- Z.S. Wang, Kevin W. Plaxco and Dmitrii E. Makarov, Influence of local, residual structure on the scaling behavior and dimensions of unfolded proteins, Biopolymers 86 (2007) 321-328 link
- Christina L. Ting and Dmitrii E. Makarov, Two-dimensional fluorescence resonance energy transfer as a probe for protein folding: A theoretical study, J. Chem. Phys. 128 (2008) 115102
link
- Reza Soheilifard, Dmitrii E. Makarov, and Gregory J. Rodin, Critical evaluation of simple network models of protein dynamics and their comparison with crystallographic B-factors, Physical Biology. 5 (2008) 026008. link
- Reza Soheilifard, Dmitrii E. Makarov, and Gregory J. Rodin, Rigorous coarse graining for the dynamics of linear systems with application to relaxation dynamics of proteins, J. Chem. Phys. 134 (2011) 054109, link
- Andrea Soranno, Brigitte Buchli, Daniel Nettels, Sonja Müller-Späth, Ryan R. Cheng, Shawn H. Pfeil, Armin Hoffmann, Everett A. Lipman, Dmitrii E. Makarov, Benjamin Schuler, Quantifying internal friction in unfolded and intrinsically disordered proteins with single molecule spectroscopy, PNAS (2012) link
- Ryan R. Cheng, Alexander T. Hawk and Dmitrii E. Makarov, Exploring the role of internal friction in the dynamics of unfolded proteins using simple polymer models, J. Chem. Phys. 138 (2013) 074112, link
- Dmitrii E. Makarov, Interplay of non-Markov and internal friction effects in the barrier crossing kinetics of biopolymers: Insights from an analytically solvable model, J. Chem. Phys. 138 (2013) 014102, link
- Ronald M. Levy, Wei Dai, Nan-Jie Deng, and Dmitrii E. Makarov, How long does it take to equilibrate the unfolded state of a protein?, Protein Science, 22, 1459-1465 (2013), doi: 10.1002/pro.2335, link
- Ignacia Echeverria, Dmitrii E. Makarov, and Garegin A. Papoian, Concerted Dihedral Rotations Give Rise to Internal Friction in Unfolded Proteins, JACS, DOI: 10.1021/ja503069k link